•
the Blog
Why do bioinformatics?
By Guillaume Filion, filed under
software pollution,
benchmark,
bioinformatics.
• 20 May 2015 •
 I never planned to do bioinformatics. It just happened because I liked to spend time in front of my computer and my boss was OK with it. Still, as every sane individual, I sometimes think that I should do something else with my life, and I wonder whether I am doing the right thing. On this topic, I recently came across the famous farewell to bioinformatics by Frederick J. Ross, which is worth reading, and from which the most emblematic quote is the now celebrated aphorism
I never planned to do bioinformatics. It just happened because I liked to spend time in front of my computer and my boss was OK with it. Still, as every sane individual, I sometimes think that I should do something else with my life, and I wonder whether I am doing the right thing. On this topic, I recently came across the famous farewell to bioinformatics by Frederick J. Ross, which is worth reading, and from which the most emblematic quote is the now celebrated aphorism
Fuck you, bioinformatics. Eat shit and die.
There is nothing to agree or disagree in this quote, but Frederick gives further detail about his point of view in the post. In short, bioinformaticians are bad programmers, and community-level obfuscation maintains the illusion.
By making the tools unusable, by inventing file format after file format, by seeking out the most brittle techniques and the slowest languages, by not publishing their algorithms and making their results impossible to replicate, the field managed to reduce its productivity by at least 90%, probably closer to 99%.
There are indeed many issues in the bioinformatics community and I am on Frederick’s side regarding file formats...
If cars were made by bioinformaticians...
By Guillaume Filion, filed under
cars,
software,
bioinformatics.
• 31 January 2015 •

1. Cars would have nice names
Here is what an abstract describing a car would look like.
Transporting people to defined locations of interest is a challenge of significant economic importance. To achieve this goal, people usually use cars or public transport. However, these solutions are suboptimal in several conditions. For instance, when people are extremely close to their target location, both cars and public transport are inappropriate, which limits their practical use. Here we present CaЯ (vehiCle for chAnging geo-cooЯdinates), a fast and accurate tool as an alternative to existing vehicles.
2. Cars would be fast and accurate
Bioinformaticians develop fast and accurate software. Their cars would be just the same. Here is what a typical benchmark sections would look like.
To show that CaЯ is faster and more accurate than existing alternatives, we benchmarked CaЯ against Volvo XC90 and Ferrari F430. In the first series of tests, we measured the time to lower the front windows of the vehicles. The average run duration was 2.3 seconds for CaЯ, 3.1 seconds for Volvo XC90 and 3.9 seconds for Ferrari F430, which demonstrates that CaЯ is substantially faster than Volvo XC90 and Ferrari F430...
Longest runs and DNA alignments 

By Guillaume Filion, filed under
sequence alignment,
BLAST,
bioinformatics.
• 31 December 2014 •
The problem of sequence alignment gets a lot of attention from bioinformaticians (the list of alignment software counts more than 200 entries). Yet, the statistical aspect of the problem is often neglected. In the post Once upon a BLAST, David Lipman explained that the breakthrough of BLAST was not a new algorithm, but the careful calibration of a heuristic by a sound statistical framework.
Inspired by this idea, I wanted to work out the probability of identifying best hits in the problem of long read alignments. Since this is a fairly general result and that it may be useful for many similar applications, I post it...
Why Linux is awesome
By Guillaume Filion, filed under
open source,
self-learning,
Linux.
• 18 December 2014 •
Before you rush to the comments and express your opinion about this title, let me make something clear. I do not expect anybody to erase their operating system and install Linux after reading this post. Actually, I do not care whether you use Linux or another operating system. All I want is to share what I have learned by using Linux, and why it made me a better scientist.
The mouse that infects your brain
 About a year after I started to use Linux, I was surprised to realize how uncomfortable working on Mac and Windows had become. I could not quite pinpoint the problem, but I had the vague feeling that something was missing. Yet everything seemed to be there. When I could finally formulate it, I realized that what was bugging me was the discomfort that all the possible options had been preconceived for me. I could click on option A, I could click on option B, and if I liked neither of those, there was no option C.
About a year after I started to use Linux, I was surprised to realize how uncomfortable working on Mac and Windows had become. I could not quite pinpoint the problem, but I had the vague feeling that something was missing. Yet everything seemed to be there. When I could finally formulate it, I realized that what was bugging me was the discomfort that all the possible options had been preconceived for me. I could click on option A, I could click on option B, and if I liked neither of those, there was no option C.
But most surprising was that I had never realized this before, because I had no idea of all the things you can do with your computer. I...
Nature on code sharing
By Guillaume Filion, filed under
open source,
reproducible research,
obfuscated open source,
bioinformatics revolution.
• 09 November 2014 •
A recent Nature editorial entitled “Code share” discusses an update in Nature’s policy regarding the use of software. Interestingly, the subtitle is
Papers in Nature journals should make computer code accessible where possible.
 Yes, finally! The last decade was a transition period, which, in the history of bioinformatics will probably be known as the “bioinformatics revolution”. Following the completion of the first genome projects, the demand for bioinformatics rose steadily, to the detriment of biochemistry and genetics, which have now fallen from grace. Something as traumatic cannot happen in a day, and it cannot happen without pain. Actually, the transition is still ongoing and this regularly causes difficulties of all kinds in biology.
Yes, finally! The last decade was a transition period, which, in the history of bioinformatics will probably be known as the “bioinformatics revolution”. Following the completion of the first genome projects, the demand for bioinformatics rose steadily, to the detriment of biochemistry and genetics, which have now fallen from grace. Something as traumatic cannot happen in a day, and it cannot happen without pain. Actually, the transition is still ongoing and this regularly causes difficulties of all kinds in biology.
One of the most perverse effects of the massive popularization of bioinformatics is that senior scientists were not properly trained for it. This led to an implicit view that bioinformatics is a tool, somewhat like a microscope or a FACS. This explains why the materials and methods section of the first papers using bioinformatics was often reduced to something like “all the bioinformatics analysis were performed using R”. In other words, “we got some bioinformatics software and asked a qualified technician to use it...
Bioinformatics without Excel
By Guillaume Filion, filed under
self-learning,
Excel,
bioinformatics.
• 27 October 2014 •
About a year after setting up my laboratory, an observation suddenly hit me. All the job applicants were biologists who wanted to do bioinformatics. I was myself trained as an experimental biologist and started bioinformatics during my post-doc. They saw in my laboratory the opportunity to do the same. Indeed, “how did you become a bioinformatician?” is a question that I hear very often.
 For lack of a better plan, most people grab a book about Linux or sign up for a Coursera class, try to do a bit every day and... well, just learn bioinformatics. I have seen extremely few people succeed this way. The content inevitably becomes too difficult, motivation decreases and other commitments take over. I will not lie, self-learning bioinformatcs is hard and it is frustrating... but it can be fun if you know how to do it. And most importantly, if you understand your worst enemy: yourself.
For lack of a better plan, most people grab a book about Linux or sign up for a Coursera class, try to do a bit every day and... well, just learn bioinformatics. I have seen extremely few people succeed this way. The content inevitably becomes too difficult, motivation decreases and other commitments take over. I will not lie, self-learning bioinformatcs is hard and it is frustrating... but it can be fun if you know how to do it. And most importantly, if you understand your worst enemy: yourself.
Here is a small digest of how it happened for me. I do not mean that this is the only way. I simply hope that this will be useful to those who seriously want to dive into bioinformatics.
Step 1. Get out of your...
A flurry of copycats on PubMed
By Guillaume Filion, filed under
PubMed,
journals,
information retrieval.
• 04 October 2014 •
It started with a search for trends on PubMed. I am not sure what I expected to find, but it was nothing like the “CISCOM meta-analyses”. Here is the story of how my colleague Lucas Carey (from Universitat Pompeu Fabra) and myself discovered a collection of disturbingly similar scientific papers, and how we got to the bottom of it.
Pattern breaker
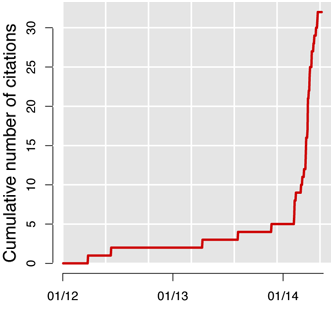 CISCOM is the medical publication database of the Research Council for Complementary Medicine. Available since 1995, it used to be mentioned in 2 to 3 papers per year, until Feburary 2014 when the number of hits started to skyrocket. Since then, “CISCOM” surfs a tsunami of one new hit per week.
CISCOM is the medical publication database of the Research Council for Complementary Medicine. Available since 1995, it used to be mentioned in 2 to 3 papers per year, until Feburary 2014 when the number of hits started to skyrocket. Since then, “CISCOM” surfs a tsunami of one new hit per week.
But this is not what drew my attention, such waves are not unheard of on PubMed. For instance, the progression of CRISPR/Cas9, is more impressive. It was the titles of the hits that convinced me that something fishy was going on: all of them are on the model “something and something else: a meta-analysis”.
The strange pattern caught my attention, but I somehow missed its significance and put this in the back of my mind. It was only later that Lucas convinced me...
(Mis)using the KS test for p-hacking 
By Guillaume Filion, filed under
statistics,
p-hacking.
• 20 September 2014 •
A colleague of mine (let’s call him John) recently put me in a difficult situation. John is a very good immunologist who, as nearly everybody in the field, had to embrace the “omics” revolution. Spirited and curious, he has taken the time to look more closely into statistics and he now has an understanding of most popular parametric and nonparametric tests. One day, he came to me with the following situation...
Shadow libraries
By Guillaume Filion, filed under
journals,
shadow libraries.
• 15 September 2014 •
 In Jorge Luis Borges’s short fiction Tlön, Uqbar, Orbis Tertius, the narrator discovers by accident the existence of a secret encyclopaedia. In this story more than in the others, Borges added many details that are actually true, in such a way that it is hard to tell the reality from the fiction. Needless to say, most would think that the secret encyclopaedia is pure fiction... and they would be wrong. Secret encyclopaedias do exist and they go by the name of “shadow libraries”.
In Jorge Luis Borges’s short fiction Tlön, Uqbar, Orbis Tertius, the narrator discovers by accident the existence of a secret encyclopaedia. In this story more than in the others, Borges added many details that are actually true, in such a way that it is hard to tell the reality from the fiction. Needless to say, most would think that the secret encyclopaedia is pure fiction... and they would be wrong. Secret encyclopaedias do exist and they go by the name of “shadow libraries”.
Academic publishing
In the course of developing PubCron (a personalized academic literature watch), I learned that PubMed references more than 2,000 new articles daily. For the vast majority of those papers, the authors pay about $1,000 as publication fees. In the bio-medical field alone, this is a $2 million gift to the publishing industry. Every day.
Gift is not the proper term. This money goes to scientific editors and in such amount, it should be sufficient to sustain about 20,000 professional editors. An editor working full time would publish 3 papers per month... not an unreasonable estimate considering that only a fraction of the manuscripts are published. This means that...
Detecting trends in culture
By Guillaume Filion, filed under
PubMed,
information retrieval,
Changepoint detection.
• 16 July 2014 •
 On June 28, 1914, Archduke Franz Ferdinand was assassinated in Sarajevo. One month later, Austria-Hungary declared war on Serbia, to which Russia responded by declaring war on Austria-Hungary, forcing its allies France and Great Britain into the war. In the aftermath, Germany honoured its defensive pact with Austria-Hungary and declared war on France, plunging Europe in a chaos that nobody had predicted.
On June 28, 1914, Archduke Franz Ferdinand was assassinated in Sarajevo. One month later, Austria-Hungary declared war on Serbia, to which Russia responded by declaring war on Austria-Hungary, forcing its allies France and Great Britain into the war. In the aftermath, Germany honoured its defensive pact with Austria-Hungary and declared war on France, plunging Europe in a chaos that nobody had predicted.
Cliodynamics, the mathematical approach to History, still has a long way to go to reach the accuracy of Isaac Asimov’s fictive psychohistory. Its closest non science-fiction relative, culturomics, relies on the idea that historical trends are accessible through the digital literature. As Jean-Baptiste Michel explains on TED, the course of History leaves a strong mark on the things we write about, and on the way we write about them.
But historical events are not the only thing we write about. The digital records are mostly about anything we find interesting. Knowing what is talked about is not science-fiction, it is actually fairly easy. More challenging is to know whether a topic is currently on the rise or merely fluctuating, which is a changepoint detection problem. Research on changepoint problems...

