A flurry of copycats on PubMed
By Guillaume Filion, filed under
PubMed,
journals,
information retrieval.
•
•
It started with a search for trends on PubMed. I am not sure what I expected to find, but it was nothing like the “CISCOM meta-analyses”. Here is the story of how my colleague Lucas Carey (from Universitat Pompeu Fabra) and myself discovered a collection of disturbingly similar scientific papers, and how we got to the bottom of it.
Pattern breaker
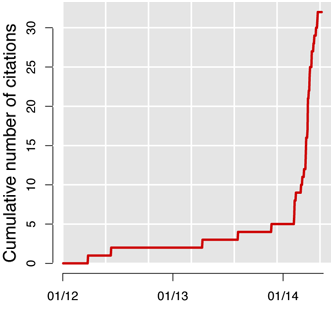 CISCOM is the medical publication database of the Research Council for Complementary Medicine. Available since 1995, it used to be mentioned in 2 to 3 papers per year, until Feburary 2014 when the number of hits started to skyrocket. Since then, “CISCOM” surfs a tsunami of one new hit per week.
CISCOM is the medical publication database of the Research Council for Complementary Medicine. Available since 1995, it used to be mentioned in 2 to 3 papers per year, until Feburary 2014 when the number of hits started to skyrocket. Since then, “CISCOM” surfs a tsunami of one new hit per week.
But this is not what drew my attention, such waves are not unheard of on PubMed. For instance, the progression of CRISPR/Cas9, is more impressive. It was the titles of the hits that convinced me that something fishy was going on: all of them are on the model “something and something else: a meta-analysis”.
The strange pattern caught my attention, but I somehow missed its significance and put this in the back of my mind. It was only later that Lucas convinced me to take a serious look at the case.
A copycat is a copycat is a copycat
We immediately noticed that the authors of all the studies are Chinese. Our first guess was that a Chinese research institute suddenly went berserk on systematic meta-analyses. We expected to find few groups of authors, or a certain geographical clustering, but there were 32 CISCOM meta-analyses, written by 28 different groups located in several cities (4 groups authored two papers). The studies are about various diseases (Crohn’s disease, coronary artery disease, cancer...) and various risk factors, going from genetic variants to aerobic exercise. There did not seem to be any special focus in the CISCOM meta-analyses (neither any obvious link with CISCOM).
The real shock came when we downloaded the papers. Most are behind paywalls, so we could access only 25 of them. There was no need for more, the pattern was very clear from the third study. The papers have exactly the same structure, with the same figures in the same order. The last figure of each paper is a “Begger’s funnel plot”, featuring a surrealistic fusion of Begg and Egger (Colin Begg and Matthias Egger both gave their name to a test for publication bias). What struck me the most was the similarity in the aesthetics of the figures, the same line widths, the same colors, the same shapes etc. As an illustration, the 8 panels below are Figure 1 taken from different papers.
It was also easy to check that some parts of the text were partly plagiarized. Below is a sentenced taken from the introduction of one of the papers.
Typically, CHD occurs when part of the smooth, elastic lining inside a coronary artery develops atherosclerosis.
And below is a passage from the Wikipedia page for coronary heart disease.
Typically, coronary artery disease occurs when part of the smooth, elastic lining inside a coronary artery (the arteries that supply blood to the heart muscle) develops atherosclerosis.
Most of the time we could not find an external source, even though the texts were very similar with each other. The discussion invariably contained the same statements in the same order and structure, with some minor variations. What do I mean by minor? Below are two example taken from different papers.
Importantly, the inclusion criteria of cases and controls were not well defined in all included studies and thus might have influenced our results.
Importantly, the inclusion criteria of cases and controls were not well defined in all included studies, which might also have influenced our results.
At that point, it was clear that the papers have not been written independently. What could make papers published by teams in different cities look so similar?
Insert title here
This question led us to take a closer look at the genealogy of the papers. We picked 13 characters, chosen to be independent of the content itself. They included annotations, figure styles, figure legends and features of the text. For instance, one of those characters is the label of the y-axis in Figure 2, coming in two flavors: “number of articles” and “number of literature”. The dendrogram of the genealogy reconstructed by maximum parsimony is shown below. The nodes of the tree represent the papers arbitrarily labelled from A to Y; an edge between them indicates that they are closest relatives.
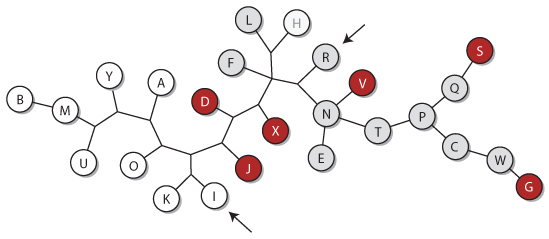
The papers in red contain the sentence “our results had lacked sufficient statistical power”, which is grammatically incorrect, and the papers in gray contain the same sentence without the erroneous had (except paper H, in which the statistical power was sufficient). The topology of the tree shows that even the most parsimonious genealogy requires 4 independent introductions of the same mistake*.
The genealogy of the papers is not consistent with the genealogy of the characters, which means that each paper derives from several templates. If there were only one template or if the writers plagiarized only one paper, the characters would coincide with the topology of the tree. Instead, it seems that the writers borrow attributes from different papers and combine them.
This makes it very unlikely that the authors wrote the papers. Most of the studies were submitted between October and November 2013, whereas the first study was published in December 2013 and indexed on PubMed in January 2014. How could the authors have plagiarized so many manuscripts before they were available? And how to explain that papers I and R (pointed on the dendrogram), signed by the same authors, are each closer to some papers signed by other authors?
And the ghostwriter is...
The only solid hypothesis is that the same ghostwriter wrote all the CISCOM meta-analyses. But how to prove or disprove it? We ventured that the ghostwriter could be a company. If this were the case, it would have to advertise its services in China, which gave us some hope to find it. With the help of a colleague in China, we found a meta-analysis writing service on the web (here is the link to their site). Calling to inquire about their services, they offered a meta-analysis published in a journal with impact factor 2-3 for around 10,000 $US. This full service cost includes writing the paper and dealing with the review process until the publication is accepted.
We do not know whether this company is the ghostwriter of the CISCOM meta-analyses, but we now know for a fact that such practice exists. The CISCOM meta-analyses may be just the tip of the iceberg. In the course of this investigation, we have come across a few similar meta-analyses not using the CISCOM database, which makes them more difficult to identify. Who knows how many papers are written by ghostwriter companies?
How to explain the high number of crossing-overs between the papers? We speculate that the authors run plagiarism checks with standard tools such as grammarly and bring minor edits until the papers pass the test, a process sometimes referred to as text laundering.
Epilogue
The work described in the CISCOM meta-analyses looks real, the literature is cited properly and is relevant to the main message... but we cannot help feeling a certain ethical unrest. We do not want to blame or judge Chinese researchers. Promotion of Chinese medical doctors is based on publications in SCI journals, which they cannot write for lack of time and training. Besides, ghost authorship in many forms is commonplace everywhere in the scientific world. Yet, the existence of ghostwriting companies rises many questions about good scientific practice. Many of those questions are also addressed to the editors publishing these studies.
More generally, this also calls for a deeper reflection about the formalism of scientific communication. In his classical paper Is the scientific paper fraudulent?, Peter Medawar wrote
The scientific paper is a fraud in the sense that it does give a totally misleading narrative of the processes of thought that go into the making of scientific discoveries.
Perhaps the time has come to explore more open ways to communicate with each other in science.