ENCODE data, Principal Components and racism
By Guillaume Filion, filed under
ENCODE,
racism,
series: genetics and racism,
Principal Component Analysis.
• 28 September 2015 •
 “Thinking is classifying” wrote Georges Clémenceau*. This tells, in simple words, everything about the obsession of the human mind to keep things tidy. No surprise we ask computers a little help here and there. Is this email spam? Is this online user human? Is this text written by that author? Training machines to put things into the boxes created by our human mind is called supervised learning and it can be very lucrative. But what about the more philosophical cases where machines make their own boxes? Can we reverse the process and put things in boxes created by computers? Unsupervised learning, as it is called, creates a lot of interesting problems where we, humans, are left wondering whether the boxes make any sense.
“Thinking is classifying” wrote Georges Clémenceau*. This tells, in simple words, everything about the obsession of the human mind to keep things tidy. No surprise we ask computers a little help here and there. Is this email spam? Is this online user human? Is this text written by that author? Training machines to put things into the boxes created by our human mind is called supervised learning and it can be very lucrative. But what about the more philosophical cases where machines make their own boxes? Can we reverse the process and put things in boxes created by computers? Unsupervised learning, as it is called, creates a lot of interesting problems where we, humans, are left wondering whether the boxes make any sense.
The mother of all classification techniques is undisputedly Principal Component Analysis (PCA). But let me reassure those who hate PCA and those who never heard of it: I will just touch the surface, and then very briefly. PCA automatically arranges similar items close to each other on a plane. The rest is up to you. Similarity, in particular, depends on a bunch of arbitrary features, size, height, number of legs... In a classical introductory example, students are asked to do a PCA on measurements of iris flowers; to their great surprise and enjoyment, the flowers gather by species. Too didactic to be true. In most real examples, beginners will watch the figure in puzzlement, like a psychic would the Tarot in search for a deep message. But admittedly, there is behind every PCA a more or less secret hope that an evident classification would spontaneously emerge from the data.
So, this bright morning I was doing a PCA on ENCODE ChIP-seq data in hope to obtain some spectacular classification. And right here is where I should reassure those who hate ENCODE and those who never heard of it... but instead I will give some background on ChIP-seq because it is more important. In a ChIP-seq experiment, every locus of a genome has a score representing the amount of time a protein spends there. Together, those scores form the profile of the protein, telling its distribution on the genome. For instance, in a ChIP-seq profile of the RNA polymerase, transcribed genes are expected to have a high score. So what I was really doing this bright morning was searching for groups of proteins with similar profiles, or similar distributions if you prefer. And this is what I got.
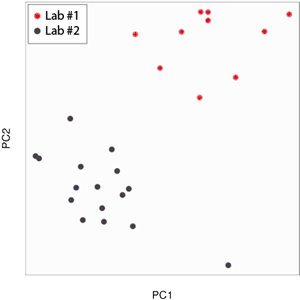
Like the iris flowers, the ChIP-seq experiments had spontaneously formed distinct groups, but the distinction had nothing to do with which protein was mapped. Instead, it only had to do with who did the experiment. This was saying that protein A in lab #1 looked more like protein B in lab #1, than protein A in lab #2. We call this “batch effect”, and this one was pretty spectacular... or so I thought.
Below is a snapshot of four profiles of the same protein**. The top two are from lab #1 and the bottom two from lab #2 (I should have included the profile of at least another protein for comparison, they look very different). It took me quite some time to find features that distinguish the top two from the bottom two profiles. Overall, my eyes told me that the reproducibility between labs was quite good. So what was going on?
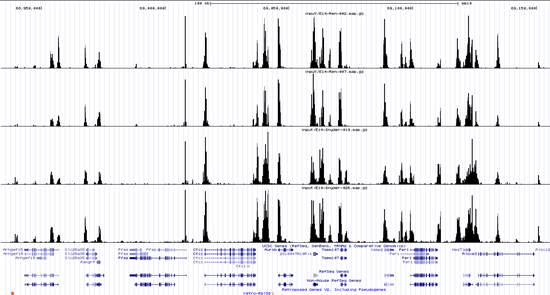
What exactly was the PCA seeing that I was not? It was tiny systematic differences scattered across the genome***. Accumulated, those differences formed a signature identifying the laboratory of origin as surely as a fingerprint. And like a fingerprint, this signature was not visible unless you specifically look for it.
The fact that experiments segregate by laboratory of origin does not mean that this dominates the signal. I will repeat this because it is important. The fact that experiments segregate by laboratory of origin does not mean that this dominates the signal. The differences between labs were tiny, but they were systematic, happening always on the same few loci. The differences between proteins were large, but unstructured, so they were not picked up by the PCA.
Understanding this is understanding a lot about the debate around the existence and the legitimacy of human races. The famous picture below is a PCA of European people arranged by genotype. The correspondence with the geographical distribution is striking. Yet, this segregation does not dominate the signal.
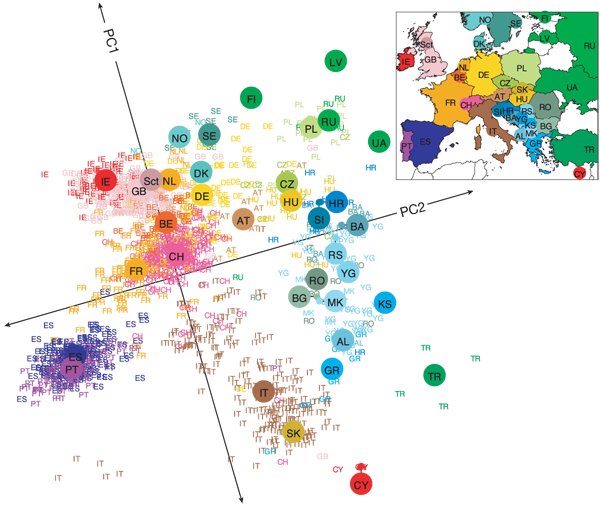
There is no doubt that we can identify the provenance of people from their genes. The question is whether races make sense, given that they tell little more than the provenance. And here, computers do not help a lot.
* The original quote taken from Au soir de la pensée (1927) is “Ainsi, connaître, penser, c’est classer”.
** It is actually the histone post-translational modification H3K4me3.
*** More specifically, the differences were at loci with high A+T content. Even though the labs were supposed to use the same protocol, I suspect that they were using different PCR polymerases.
« Previous Post | Next Post »
blog comments powered by Disqus

