Who understands the histone code?
By Guillaume Filion, filed under
histone code,
position effect variegation,
epigenetics.
• 30 November 2013 •
The most annoying thing about us biologists is that we keep using words that we don’t understand. “Epigenetics” is one of those that has drawn my attention for several years, as I explained in my last post. I suggested that the invasion started in 2001, the year that the histone code hypothesis was proposed by Thomas Jenuwein and David Allis in a seminal paper entitled Translating the Histone Code.
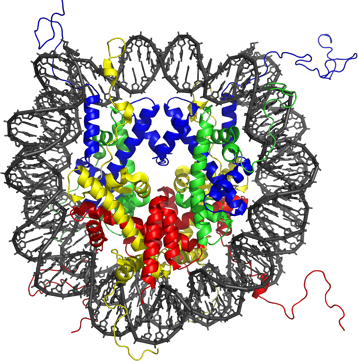 The histone code hypothesis was arguably the most influential concept of the last decade in molecular biology. Yet, most biologists would be hard pressed to say what the hypothesis is. All you have to do is read what Thomas Jenuwein and David Allis actually wrote. But believe it or not, this blog is one of the only places on the Internet where the histone code hypothesis is spelled out clearly. Most sources, including the Wikipedia article diverge substantially from the original statement, which is the following.
The histone code hypothesis was arguably the most influential concept of the last decade in molecular biology. Yet, most biologists would be hard pressed to say what the hypothesis is. All you have to do is read what Thomas Jenuwein and David Allis actually wrote. But believe it or not, this blog is one of the only places on the Internet where the histone code hypothesis is spelled out clearly. Most sources, including the Wikipedia article diverge substantially from the original statement, which is the following.
Distinct qualities of higher order chromatin, such as euchromatic or heterochromatic domains, are largely dependent on the local concentration and combination of differentially modified nucleosomes.
DNA in the nucleus comes in a structure called the nucleosome. The picture above is a molecular model of the nucleosome, showing the DNA (in black) wrapped around histones (in colors). The nucleus of a human cell contains about 10 million of those, and according to the histone code hypothesis, their different chemical flavors is what structures the genome at a larger scale. But what does it have to do with epigenetics?
The term “epigenetic” appears casually without definition throughout Translating the Histone Code, as if everybody expected to see it there. I don’t know whether the connection between “epigenetics” and “histones” was obvious for everybody at the time, but for Thomas Jenuwein it was. One year before the publication of the histone code hypothesis, he had solved one of the oldest puzzles of genetics.
The puzzle in the eye
I like to think that modern genetics started a good morning of 1910. That day Thomas Hunt Morgan noticed that a fly in his stock was looking different from the others. Instead of the usual bright red, its eye was completely white. Working from there, he realized that the mutation was heritable and his follow-up experiments led to the conclusion that genes must lie on the chromosomes.
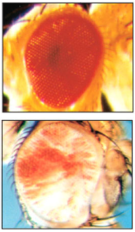 All the subsequent observations were consistent with the chromosomal theory of heredity, until twenty years later Hermann Joseph Müller mutagenized fruit flies by X-rays. After crossing a mutant that had a notch in the wing with Morgan’s white mutant, he obtained one of the most beautiful and intriguing monsters of genetics. The eye of the mutant was neither red nor white, it was red and white. The picture on the left shows the head of a fruit fly with a normal eye, above the head of Müller’s mutant with its mottled red and white patches. The significance of this discovery did not escape Müller. He fully understood that this variation in a single individual was challenging the dogma. How could a fly have the mutation and not have it at the same time? His explanation was that the mutation was flickering on and off, or “eversporting” as he put it himself in a seminal paper.
All the subsequent observations were consistent with the chromosomal theory of heredity, until twenty years later Hermann Joseph Müller mutagenized fruit flies by X-rays. After crossing a mutant that had a notch in the wing with Morgan’s white mutant, he obtained one of the most beautiful and intriguing monsters of genetics. The eye of the mutant was neither red nor white, it was red and white. The picture on the left shows the head of a fruit fly with a normal eye, above the head of Müller’s mutant with its mottled red and white patches. The significance of this discovery did not escape Müller. He fully understood that this variation in a single individual was challenging the dogma. How could a fly have the mutation and not have it at the same time? His explanation was that the mutation was flickering on and off, or “eversporting” as he put it himself in a seminal paper.
The chromosome or gene controlling the eye colour in this case must be subject to frequent genetic changes during eye development, ie must somehow be “eversporting”, somewhat like the genes for variegated pigmentation in corn and in some other plants.
Müller knew that the genetic defect of this mutant was that a piece of the X chromosome carrying the white gene was translocated near the centromere of chromosome 3. Similarly, in all the mottled mutants he ever obtained, the white gene was at a new position of the genome, explaining why the phenomenon became known as Position Effect Variegation.
In spite of the initial enthusiasm, progress towards understanding Position Effect Variegation was slow. Over the years, several suppressor mutations (making the eye almost completely red), collectively called “suppressors of variegation” were isolated. Yet, they turned out to be impossible to map and identify until molecular genetics made significant progress in the 1990’s, which is where we left Thomas Jenuwein.
Su(var)3-9, BLAST and RuBisCO
The mutant that lifted the veil on Position Effect Variegation was called Su(var)3-9, for “suppressor of variegation 3-9”, which was mapped and sequenced in 1994 by Gunter Reuter. But you cannot read the function of a gene in its sequence, so the holy grail was to understand what these genes do to cause randomness in the expression of the white gene.
Somewhat unexpectedly, the answer came from bio-informatics. The BLAST software was up and running since 1990, but it was only when the Internet came of age and when online servers were available that it became used by lay biologists. A BLAST search of Su(var)3-9 against all gene products revealed a striking similarity with RuBisCO, the key enzyme of photosynthesis. More specifically, the similarity was restricted to a protein-methyltransferase domain of RuBisCO, which suggested that Su(var)3-9 encodes a protein-methyltransferase (an enzyme that transfers methyl groups on proteins).
It could have taken forever to identify the proteins methylated by Su(var)3-9, if the answer had not been suggested by previous work in neighboring fields. I will simply say that the work on two other genes, MeCP2 and Rpd3 had shown that chemical modifications of histones had an impact for the expression of genes. Thomas Jenuwein and his team thought that Su(var)3-9 encoded an enzyme that can transfer methyl groups on histones, and they were right. Methylation of the histones is what drives Position Effect Variegation.
Closing the loop
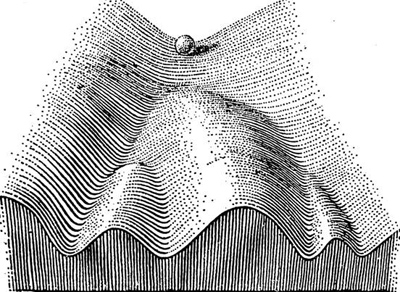 This long journey took us from histone modifications to Position Effect Variegation and back, but I still did not say anything about epigenetics. Müller’s white mottled mutant belongs to a category of developmental phenomena that contradict or extend Mendelian genetics (other classical examples include X inactivation and parental imprinting). It is therefore a true epigenetic phenomenon in the sense of Conrad Waddington, who defined epigenetics in 1940 as “the branch of biology which studies the causal interactions between genes and their products which bring the phenotype into being”. Portrayed above is his famous epigenetic landscape, which depicts biological development as the trajectory of a ball down a slope with different valleys. Genes determine the landscape, but the path of the ball has a random component.
This long journey took us from histone modifications to Position Effect Variegation and back, but I still did not say anything about epigenetics. Müller’s white mottled mutant belongs to a category of developmental phenomena that contradict or extend Mendelian genetics (other classical examples include X inactivation and parental imprinting). It is therefore a true epigenetic phenomenon in the sense of Conrad Waddington, who defined epigenetics in 1940 as “the branch of biology which studies the causal interactions between genes and their products which bring the phenotype into being”. Portrayed above is his famous epigenetic landscape, which depicts biological development as the trajectory of a ball down a slope with different valleys. Genes determine the landscape, but the path of the ball has a random component.
Over the years, the meaning of “epigenetics” progressively started to designate DNA sequence-independent heredity, but the term nevertheless remained associated to Position Effect Variegation. This is how I interpret the recurrent association between histone modifications and epigenetics in Translating the Histone Code. To my knowledge, histone modifications were never proven to be epigenetic in the modern sense, so the confusion between epigenetics and histone modifications is somewhat of a misunderstanding.
By the way, you may wonder whether the histone code hypothesis is correct or not. Data obtained by chromatin IP show a very good agreement between the nature of the genomic domains and the chemical modifications of the nucleosomes, but those modifications may as well be a consequence of the organization rather than a cause. So, as stated above, the question is still unresolved.
« Previous Post | Next Post »
blog comments powered by Disqus

