What’s in a title?
By Guillaume Filion, filed under
PubMed,
journals,
information retrieval.
• 10 March 2012 •
Trying to come up with a name for the blog, I wondered what a good title should be. If you ever wrote a scientific article, you probably found yourself in the same situation. You try to surf the trend, mix in carefully selected buzzwords and present the work under its sexiest side. Sexy, that is, to the veterans. Admittedly, not everyone will crave to read “Epithelial cell adhesion molecule (EpCAM) complex proteins promote transcription factor-mediated pluripotency reprogramming” (no offense intended, I just took the first title that showed up in PubMed).
Meta-analysis of scientific literature tells us a lot about how science and scientific discourse change over time. A simple title word analysis of the articles published in Nature in an 8 year interval shows how some topics fell from grace, whereas others rose to the top.
The struggle-for-hype allows us to tell what scientists and editors find exciting at a given time. To play with this idea, I collected all the titles of the Nature articles published in 2002 and in 2010, and ran Wordle on them. The size of a word in the cloud is proportional to its occurrence in the corpus, giving an immediate feel for which topics are the hottest.
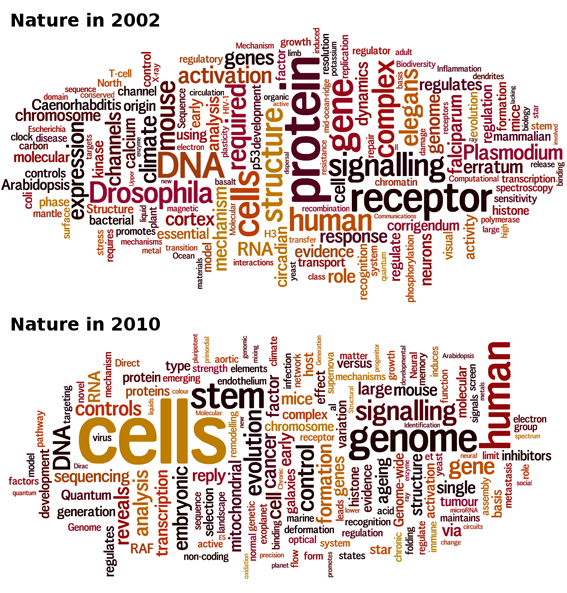
At a glance, it is obvious that “protein”, “receptor”, and “Drosophila” fell into oblivion. Staring a little longer, you will notice that “Arabidopsis”, “Caenorhabditis”, “elegans”, “Escherichia”, “coli” and “Plasmodium” also left the house and that “stem" and “cell” recently came into light.
OK, stem cells are hotter than protein receptors, so be it. But what happened to model organisms? What happened to flies (Drosophila melanogaster), worms (Caenorhabditis elegans), plants (Arabidopsis thaliana) an non mammals in general (Escherichia coli is a bacterium and Plasmodium is the malaria parasite)? Only yeast maintains itself at a low but steady level. Is that a whim of fashion or are model organisms truly on the decline? Well, experience shows that microorganisms with a glorious past such as phage lambda or E. coli are gone for good. It seems the mammalian toolbox is now mature enough to take over the niche. To blame is probably the discovery of RNA interference, which opened unlimited possibilities of manipulating gene expression and tempered the utility of mutant collections.
This analysis also says something about how scientists talk about their work. The word “reveals” seems to appear out of nowhere. Did research papers in 2002 not reveal anything? Difficult to believe. Overall, “reveals” has even been declining in the English language. Actually, “reveals” is the trademark of high-throughput research. Big data factories find their way to Nature publications more often than small labs. Large datasets lend themselves to observations and typically “reveal” something, more than they “show”, “suggest” or “prove” it. All in all, “reveals” shows that observational studies have dominated the last years.
The clouds capture many other trends, but most surprising to me is the utter disappearance of “erratum” and “corrigendum”. Would Nature have become a serious journal in the end?
If you try it in your browser, the result will look funny, you need to check the source of the page, which contains the XML data. The good news is that all the titles are there in the field
« Previous Post | Next Post »
blog comments powered by Disqus

